QSAR, ADMET & Predictive Toxicology
Tools for Designing Therapeutics with Favorable Pharmacokinetic Properties and Safety Profiles
Discover Safe and Effective Therapeutics
Understanding and quantifying structure-activity relationships can significantly impact lead optimization and drug development by minimizing tedious and costly experimentation. Build, validate and apply your own models based on a wide range of approaches, and keep improving them as new data become available. Assess the potential risk posed by unfavorable pharmacokinetic properties and potential toxicity using BIOVIA Discovery Studio’s distributed models, extend them to better cover your proprietary chemical space and use the comprehensible indications to navigate around such liabilities.
- QSAR
- ADMET
- Toxicity
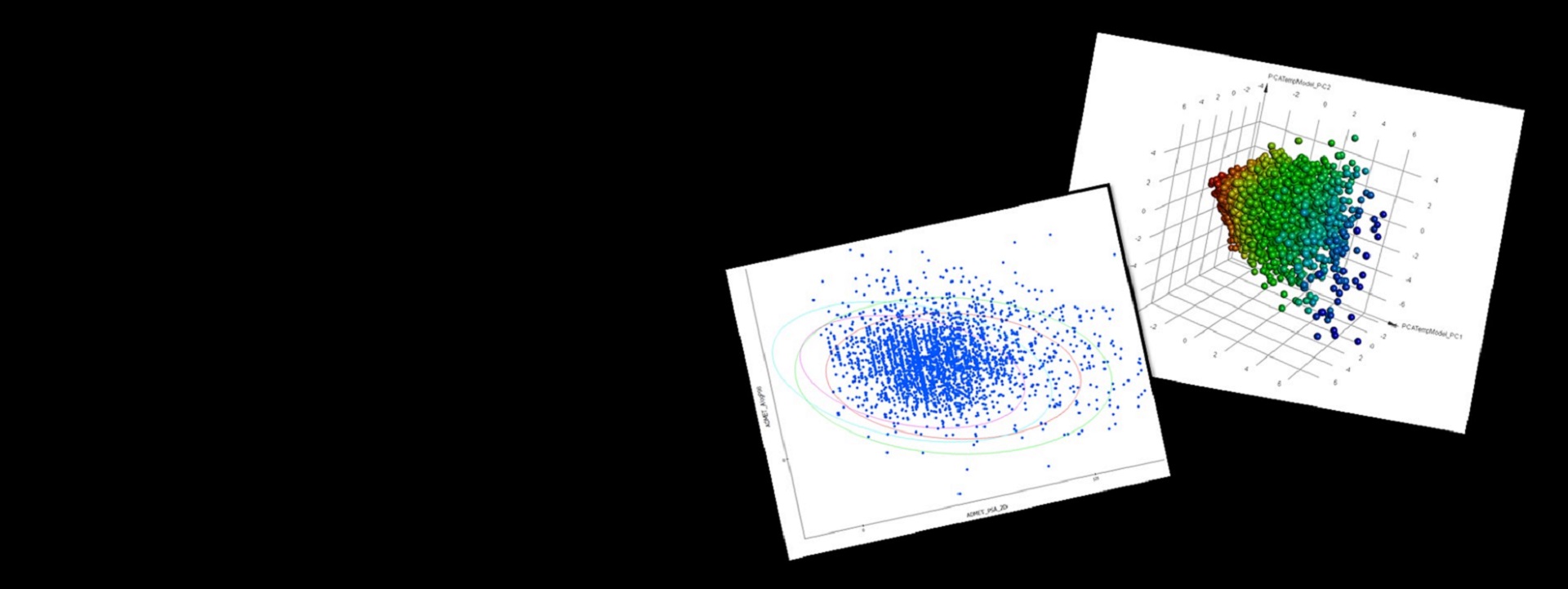
QSAR
- Comprehensive and consistent data preparation:
- Ligands: remove duplicates and handle tautomers and ionization
- Prepare response property (scaling and binning)
- Split your data into training and test sets with the appropriate methods
- Choose from a large number of physicochemical, topological, electronic, geometric. fingerprint and Quantum Mechanics based descriptors
- Create statistical models including Bayesian, MLR (Multiple Linear Regression), PLS (Partial Least Squares), and GFA (Genetic Functional Analysis)
- Analyze and validate models using model applicability domains (MAD), automatic test set validation, cross validation and statistical metrics
- Identify Matched Molecular Pairs (MMPs) transformations and study activity cliffs
ADMET
Get an early assessment of your compounds by calculating the predicted ADMET (absorption, distribution, metabolism, excretion and toxicity) properties for collections of molecules such as synthesis candidates, vendor libraries, and screening collections. Use the results to eliminate compounds with unfavorable ADMET characteristics and evaluate proposed structural refinements, designed to improve these properties prior to synthesis.
ADMET descriptors include:
- Human intestinal absorption
- Aqueous solubility
- Blood brain barrier penetration
- Plasma protein binding
- CYP2D6 binding
- Hepatotoxicity
- Filter sets of small molecules for undesirable function groups based on published SMARTS rules
Toxicity
Evaluate your compounds’ performance in experimental assays and animal models. Compute and validate assessments of the toxic and environmental effects of chemicals solely from their molecular structure. TOPKAT® (TOxicity Prediction by Komputer Assisted Technology) employs robust and cross-validated Quantitative Structure Toxicity Relationship (QSTR) models for assessing various endpoints and utilizing the patented Optimal Predictive Space validation method to assist in interpreting the results.
Please note: details on our extensible TopKat models have been published in QSAR Model Report Format (QMRF) on the European Commission Joint Research Center (JRC)’s "QSAR Model Database".
- Ames mutagenicity
- Rodent carcinogenicity (NTP and FDA data)
- Weight of evidence carcinogenicity
- Carcinogenic potency TD50
- Developmental toxicity potential
- Rat oral LD50
- Rat maximum tolerated dose
- Rat inhalation toxicity LC50
- Rat chronic LOAEL
- Skin irritancy and sensitization
- Eye irritancy
- Aerobic biodegradability
- Fathead minnow LC50
- Daphnia magna EC50
Start Your Journey
The world of QSAR, ADMET & Predictive Toxicology is changing. Discover how to stay a step ahead with BIOVIA.
Join the conversation in the BIOVIA Drug Discovery & Development Community!
Also Discover
Learn What BIOVIA Can Do for You
Speak with a BIOVIA expert to learn how our solutions enable seamless collaboration and sustainable innovation at organizations of every size.
Get Started
Courses and classes are available for students, academia, professionals and companies. Find the right BIOVIA training for you.
Get Help
Find information on software & hardware certification, software downloads, user documentation, support contact and services offering