Ligand- and Pharmacophore-based Design
Enhance Small Molecule Therapeutics Design with Best-in-Class Tools
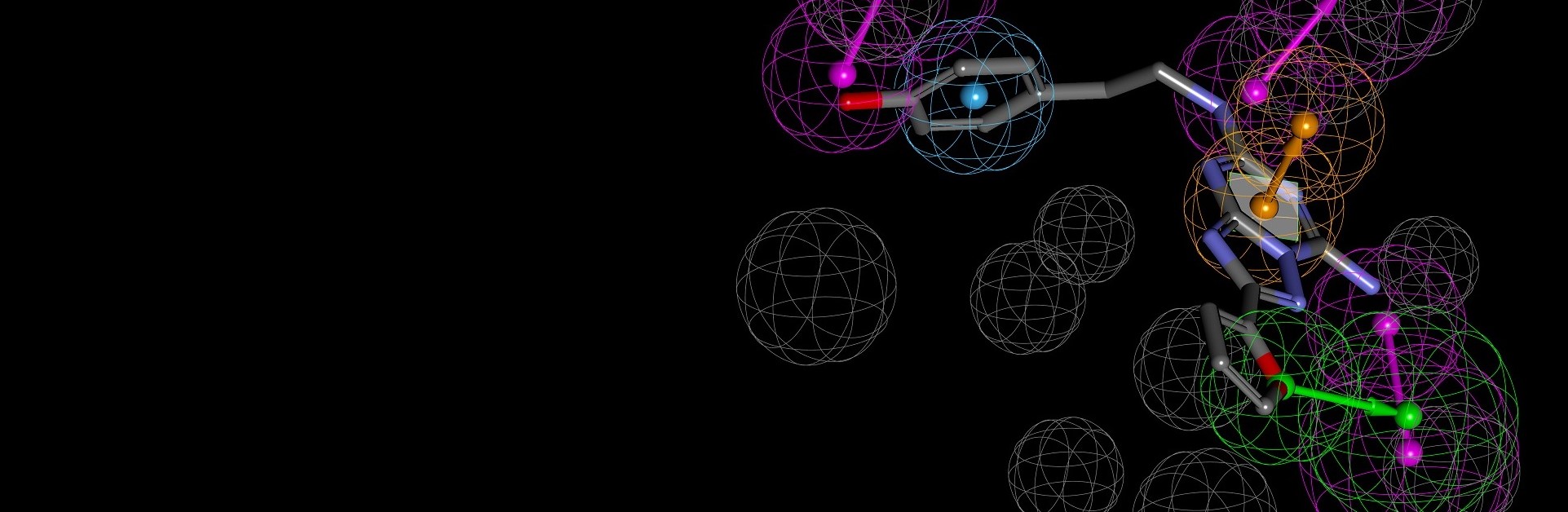
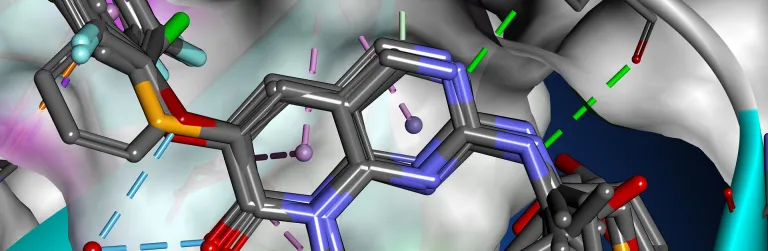
The binding interactions of proteins and ligands arise from a variety of steric and electrochemical factors. Understanding these interactions can help researchers more rapidly identify promising new therapeutic candidates. Pharmacophores map these features in 3D space, providing a simple means to screen libraries of compounds virtually, characterize the binding of existing leads and optimize their performance.
BIOVIA Discovery Studio utilizes the CATALYST Pharmacophore Modeling and Analysis toolset to assist in the assessment of small molecule therapeutics with or without target-structured data. It supports de novo drug design, multi-target drug design and activity profiling to drive small molecule R&D.
- Build
- Apply
- Design
Build
- Automatically generate pharmacophores from the data available
- Sets of active ligands
- Receptor binding sites
- Receptor-ligand complexes
- Perform rigorous Pharmacophore validation based on sets of control compounds with known activity.
- Hypotheses can include
- Geometric, feature-based queries
- Shape-similarity
- “Forbidden” space
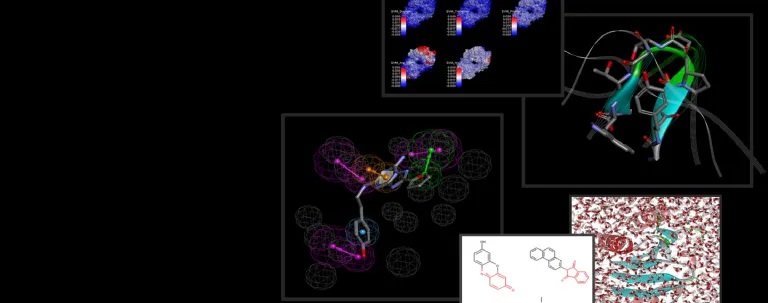
- Go beyond the limitations of classical pharmacophore elucidation algorithms by exploring Ensemble Pharmacophores for very large/diverse compound sets with a risk of multiple modes of action
Apply
Conduct robust Pharmacophore screening studies
- Build and search databases of 3D conformations
- Consider and analyze the full conformational space of your ligands
- Explore off-target activity and drug repurposing using the PharmaDB* database
Discovery Studio now includes the most extensive reported database for ligand profiling. Built from, and validated using, the scPDB*, the PharmaDB contains approximately 240,000 receptor-ligand pharmacophore models.
Design
Design and characterize your ligands and combinatorial libraries
- Enumerate reaction- or core-based libraries
- Enumerate ionization states, tautomers and isomers
- Filter poor candidates with undesirable functional groups and Lipinski and Veber rules or your own criteria
- Calculate numerous physicochemical and fingerprint properties
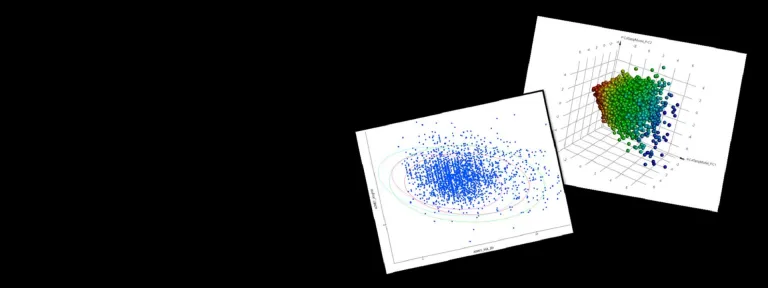
- Optimize combinatorial libraries using Pareto optimization, diversity and similarity analysis
- Clustering tools and 3D visualization using PCA analysis
Start Your Journey
Accelerate your drug discovery with BIOVIA Discovery Studio.
Join the conversation in the BIOVIA Drug Discovery & Development Community!
Also Discover
Learn What BIOVIA Can Do for You
Speak with a BIOVIA expert to learn how our solutions enable seamless collaboration and sustainable innovation at organizations of every size.
Get Started
Courses and classes are available for students, academia, professionals and companies. Find the right BIOVIA training for you.
Get Help
Find information on software & hardware certification, software downloads, user documentation, support contact and services offering