Macromolecule Design and Analysis
Comprehensive Portfolio of Validated Scientific Tools for Every Aspect of Macromolecule-based Research
Enhance Macromolecule-Based Research
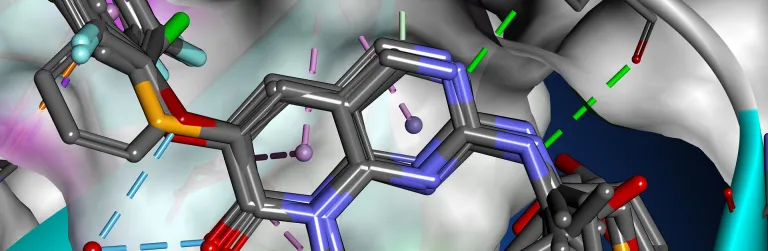
Determining the three-dimensional structure and properties of macromolecules, such as enzymes and antibodies, is a fundamental component to a wide range of research activities. For example, different conformations which arise from normal molecular dynamics or interactions with ligands or other proteins may reveal novel binding sites or provide clues as to its function. While thousands of molecules have had their structures resolved experimentally, obtaining high fidelity structural data remains a nontrivial process.
Simulation can augment physical experimentation by providing insight into macromolecular structure. Additionally, techniques such as homology modeling can help predict structural models for novel molecules, guiding therapeutic design and protein engineering efforts. BIOVIA Discovery Studio delivers a comprehensive portfolio of market leading, validated scientific tools, able to assist in every aspect of macromolecule-based research.
- Search
- Model
- Design
Search
- Perform multiple sequence searches using BLAST and PSI-BLAST against local or NCBI databases
- For multiple chain proteins, simultaneously and independently perform multiple sequence alignments of each protein chain
- Predict the transmembrane helices in transmembrane protein sequences
- Predict sites prone to Post Translational Modifications (PTMs) using sequence-based motif searching
Model
- Analyze and prepare structures from 3D structure repositories (e.g., PDB)
- Generate 3D structure models using MODELER
- Verify the quality of a structure model
- Use LOOPER to systematically search loop conformations and rank using CHARMm
- Graft loop conformations from a template structure onto a target model
- Systematically optimize amino acid side-chains using ChiRotor CHARMm simulations
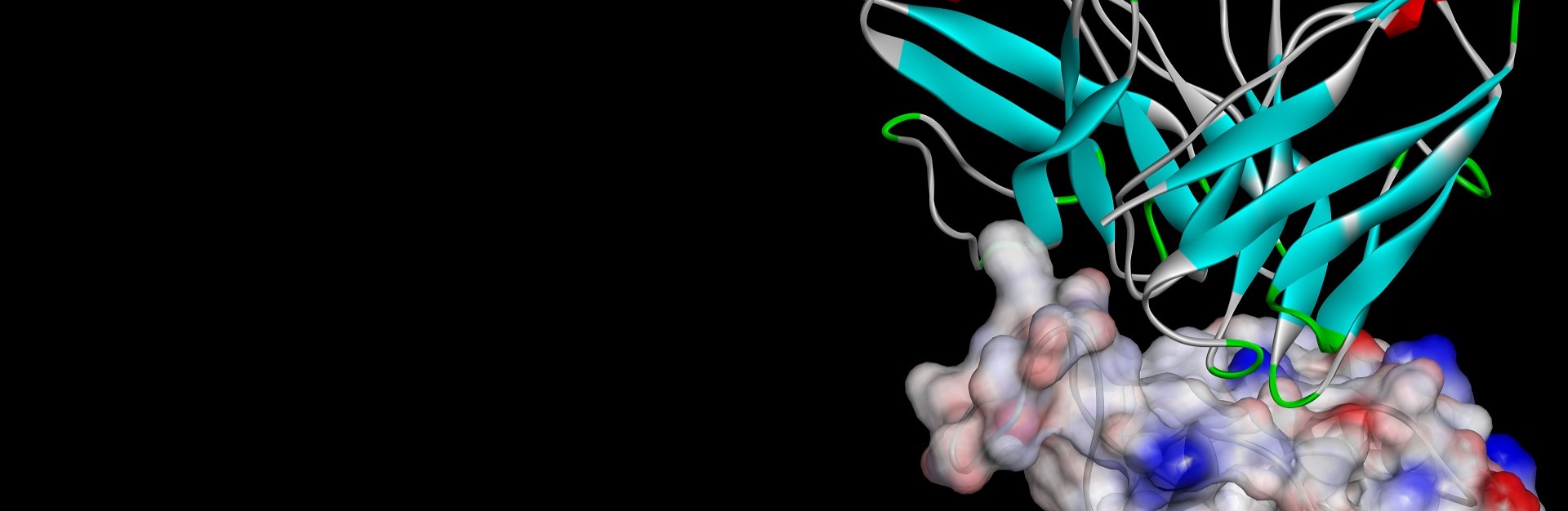
- Use ZDOCK to perform protein-protein docking and examine binding partner interactions
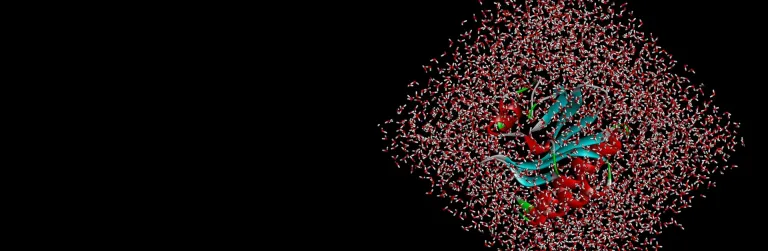
- Study conformational flexibility with explicit solvent-based Molecular Dynamics (MD) simulations using CHARMm or NAMD
Design
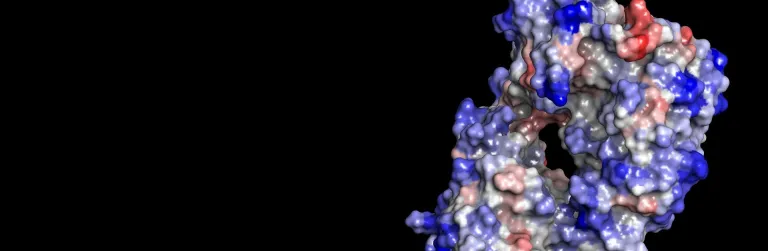
- Predict electrical properties of the protein, including pH-dependent stability and protonation states and the isoelectric point
- Conduct thermal or pH-based mutational stability and binding affinity predictions
- Identify potential stable disulfide bridge locations
- Calculate biophysical properties important for protein formulation, including viscosity and solubility
Start Your Journey
Accelerate your drug discovery with BIOVIA Discovery Studio.
Join the conversation in the BIOVIA Drug Discovery & Development Community!
Also Discover
Learn What BIOVIA Can Do for You
Speak with a BIOVIA expert to learn how our solutions enable seamless collaboration and sustainable innovation at organizations of every size.
Get Started
Courses and classes are available for students, academia, professionals and companies. Find the right BIOVIA training for you.
Get Help
Find information on software & hardware certification, software downloads, user documentation, support contact and services offering