Biotherapeutics and Antibody Modeling
A Rich Set of In Silico Tools to Support The Design of Biotherapeutics
Enhance Biotherapeutics Design and Development
Biotherapeutics offer a range of unique benefits over small molecule drugs. Their increased affinity and selectivity have allowed researchers to address new and difficult targets. This has led to a dramatic increase in R&D projects centered on antibodies and other biological modalities for treatment such as bispecifics. However, these projects must also overcome similar challenges in safety and pharmacokinetics faced by small molecules, albeit from a different perspective. Factors such as high immunogenicity or low solubility can kill development on an otherwise effective candidate.
Modeling and simulation can significantly advance the development of these new classes of therapeutics. For example, researchers can predict an antibody’s formulation properties and suggest mutations to improve them rapidly and at low cost compared to laboratory experimentation alone. BIOVIA Discovery Studio offers a rich set of tools to help guide the design of biotherapeutics, allowing teams to optimize the performance of candidates in silico and streamlining their physical work.
- Model
- Design
- Predict
Model
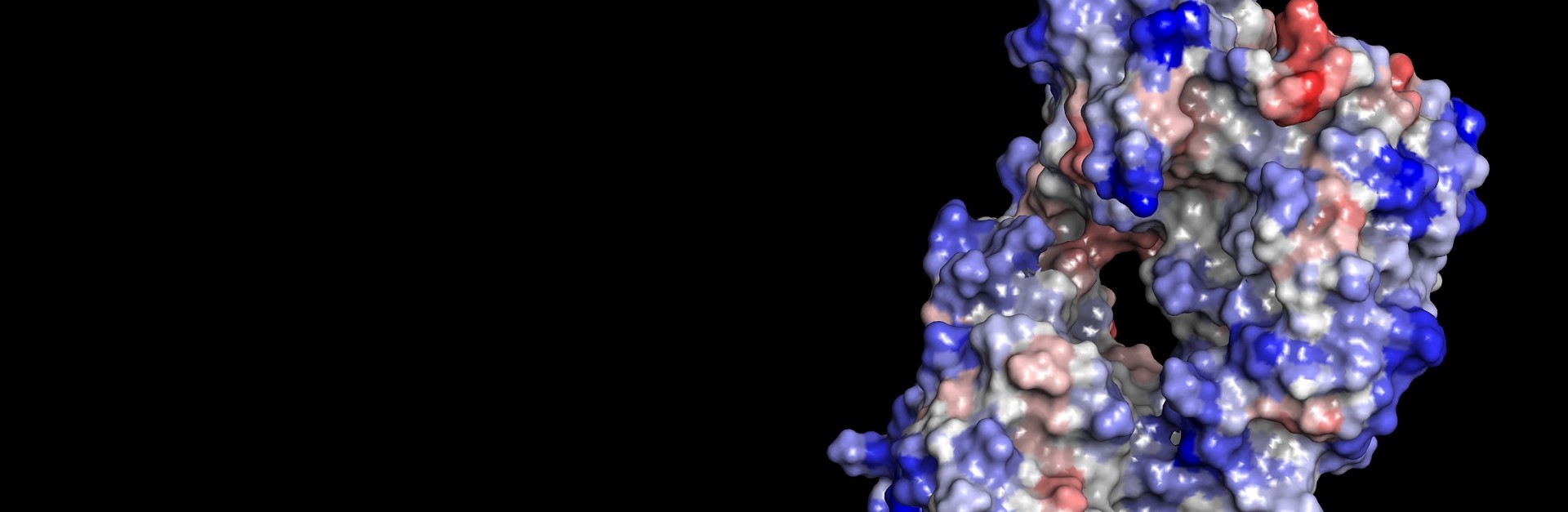
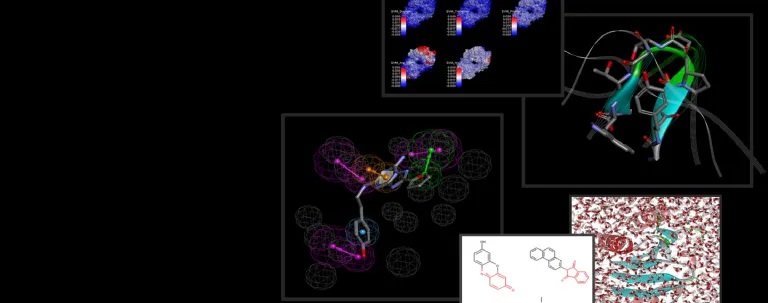
- Automated modeling cascade to easily and rapidly generate high quality 3D antibody full-length, Fab or Fv models from a set of light and heavy antibody chain sequences and a curated PDB antibody template database
- Full support for commonly used antibody annotation schemes - IMGT, Chothia, Kabat and Honegger
- Simultaneously and independently perform multiple sequence alignments of heavy and light chains
- Additional expert tools to identify antibody template structures and build high quality homology models
- Ability to build bispecific full length antibody structures
- Refine CDR loops using either template, loop grafting or de novo loop modeling
- Perform detailed model analysis
Design
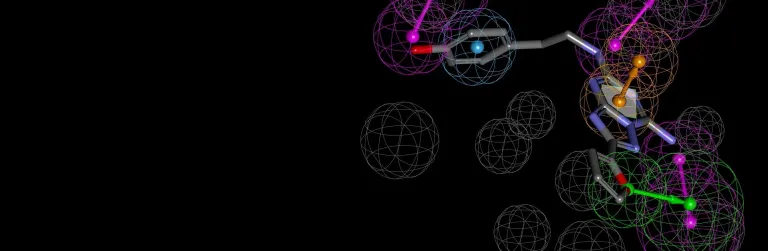
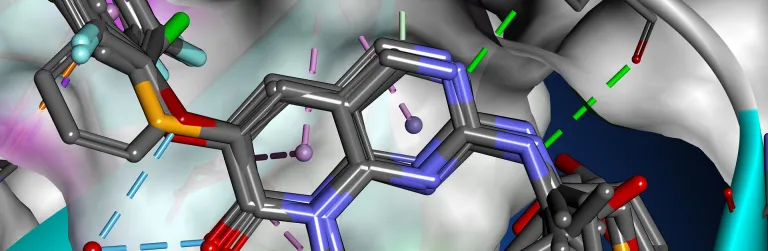
- Conduct thermal or pH-based mutational stability and binding affinity predictions
- Identify potential stable disulfide bridge locations
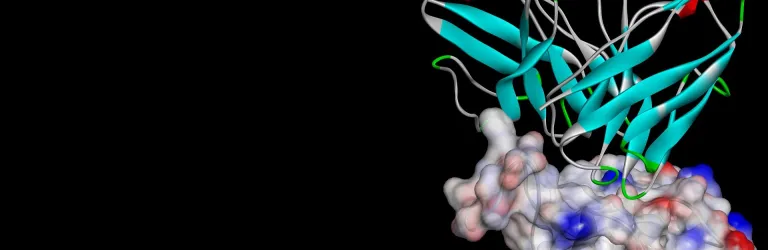
- Perform antibody-antigen docking using ZDOCK to identify critical interacting residues
- Predict residue mutations for antibody humanization without compromising antibody stability or efficacy
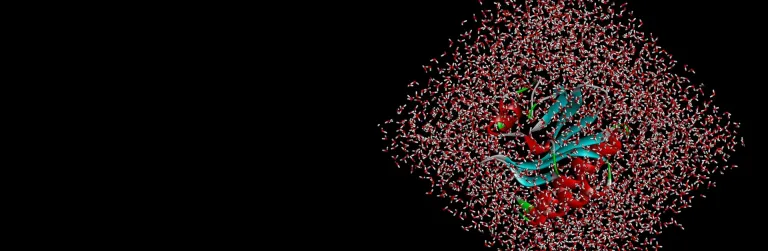
- Study conformational flexibility with explicit solvent-based Molecular Dynamics (MD) simulations using CHARMm or NAMD
Predict
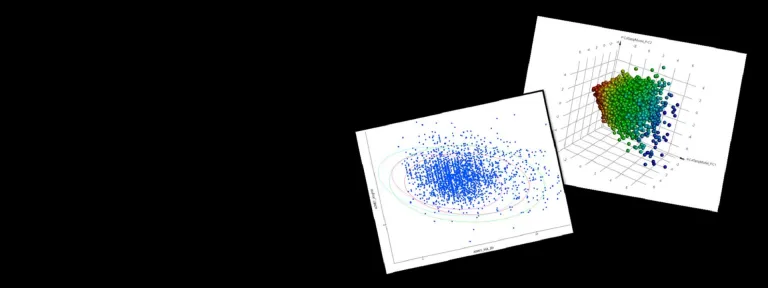
- Calculate biophysical properties including isoelectric point, solubility, viscosity (SCM*) and aggregation propensity (Developability Index – SAP*) for early stage assessment of suitability for development
- Predict sites prone to Post Translational Modifications (PTMs) using sequence-based motif searching
*In-licensed from MIT
Start Your Journey
The world of Biotherapeutics and Antibody Modeling is changing. Discover how to stay a step ahead with BIOVIA
Join the conversation in the BIOVIA Drug Discovery & Development Community!
Also Discover
Learn What BIOVIA Can Do for You
Speak with a BIOVIA expert to learn how our solutions enable seamless collaboration and sustainable innovation at organizations of every size.
Get Started
Courses and classes are available for students, academia, professionals and companies. Find the right BIOVIA training for you.
Get Help
Find information on software & hardware certification, software downloads, user documentation, support contact and services offering